SMBLImporter.jl
SBMLImporter.jl is a Julia importer for ODE models specified in the Systems Biology Markup Language (SBML). It supports many SBML features, such as events, rate-, assignment-, algebraic-rules, dynamic compartment size, and conversion factors.
Availability: https://github.com/sebapersson/SBMLImporter.jl
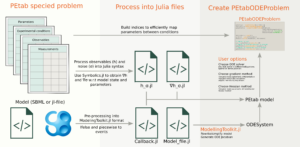
PEtab.jl
Julia package that imports ODE parameter estimation problems specified in the PEtab format. PEtab.jl uses Julia’s DifferentialEquations.jl package for ODE solvers and ModelingToolkit.jl for symbolic model processing, which enables fast model simulations combined. Additionally, it supports gradients using both forward and adjoint sensitivity approaches and Hessians via exact and approximate methods, making it suitable for efficient parameter estimation for models of various sizes. In an extensive benchmark, PEtab.jl was frequently 2-4 times faster than the pyPESTO toolbox, which utilizes the AMICI interface to the Sundials suite.
Availability: https://github.com/cvijoviclab/PEtab.jl
PEPSDI (Particles Engine for Population Stochastic DynamIcs) framework
Bayesian inference framework for dynamic state space mixed effects models (SSMEM).
Availability: https://github.com/cvijoviclab/PEPSDI

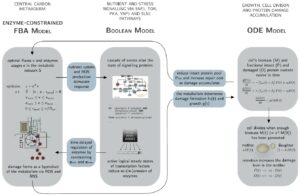
yMSA (Multi-Scale Model of Yeast Replicative Ageing)
The model consists of three interconnected modules: a Boolean signalling network model, an enzyme-constrained flux balance model of the central carbon metabolism, and a dynamic model of growth and protein damage accumulation with discrete cell divisions.
Availability: https://github.com/cvijoviclab/IntegratedModelMetabolismAgeing
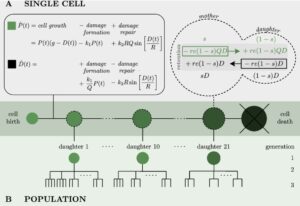
Single-cell and whole population model of damage accumulation in yeast
A dynamic model based on ordinary differential equations (ODE) capable of simulating and mapping replicative ageing on single-cell and population level. The model includes novel features on the population level, such as the health span and the rejuvenation index, in addition to previously known metrics, such as the replicative life span, growth per cell cycle and cumulative growth, characterising the precise large-scale impact of single-cell properties on the lineage.
Availability: https://github.com/cvijoviclab/RejuvenationProject
Yeast Hybrid Model
A hybrid model, combining a Boolean module, describing the main pathways of glucose and nitrogen signalling, and an enzyme-constrained model accounting for the central carbon metabolism of Saccharomyces cerevisiae, using a regulatory network as a link.
Availability: https://github.com/cvijoviclab/YeastHybridModelingFramework/
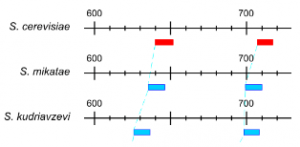
Automated heuristic detection of upstream Open Reading Frames in yeast
A series of Perl scripts for large-scale analysis of yeast 5’UTR.
Availability: http://www.molbio.gu.se/per/web/